Publications
Our scientists regularly publish in their fields' most influential peer-reviewed journals. Here is a selection of our contributions.
The Proprietary AI platforms at Deep Genomics consist of datasets, data processing pipelines, machine learning systems, including foundation models and large language models, and software engineering systems, plus the processes and protocols followed by team members.
In the AI community, a foundation model is a very large machine learning model that can be used for a wide range of tasks. In drug discovery, a traditional AI model may be good at one task, such as predicting molecule-target interactions, whereas a foundation model will have learned fundamental aspects of biology and chemistry that benefit many tasks.
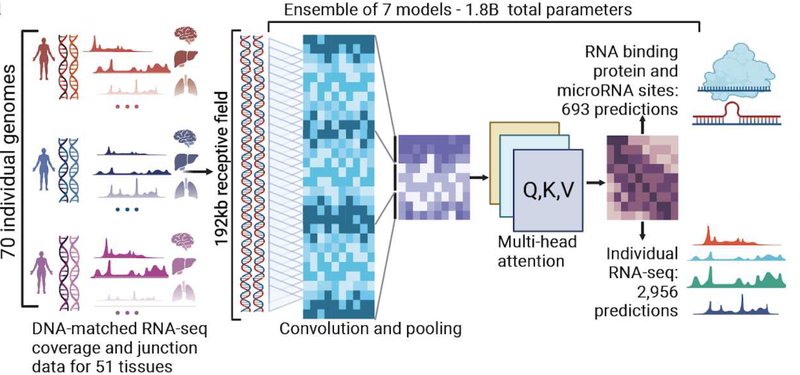
This means that BigRNA can uniquely discover a wide range of new biological mechanisms and RNA therapeutic candidates that would not be found using traditional approaches. We have built, and continue to improve upon, our proprietary BigRNA platform, which is fueled by diverse proprietary datasets, new ML engineering advances, and the ongoing work of our scientists.
Currently, most efforts have focused on predicting data that measures overall gene expression levels, which are not suited to predicting regulatory interventions; for example, specific transcriptional perturbations on splicing or polyadenylation. In contrast, BigRNA is trained to predict RNA expression at sub-gene resolution.
Our BigRNA model is good at target identification, discovering novel biological mechanisms that can be drugged, predicting molecule-target interactions, designing therapeutic candidates, designing surrogate molecules for in vivo testing, and much more. Further, it can do all of this across a wide range of species, tissues, cell models, and RNA therapeutic modalities, including oligonucleotides, DNA editing, RNA editing, and mRNA. For some of these tasks, there are individual tools, such as Enformer, Saluki, or SpliceAI, but we found that BigRNA exceeds the state of the art across a wide range of tasks.
Our scientists regularly publish in their fields' most influential peer-reviewed journals. Here is a selection of our contributions.