Daniele Merico, Head of Target Identification
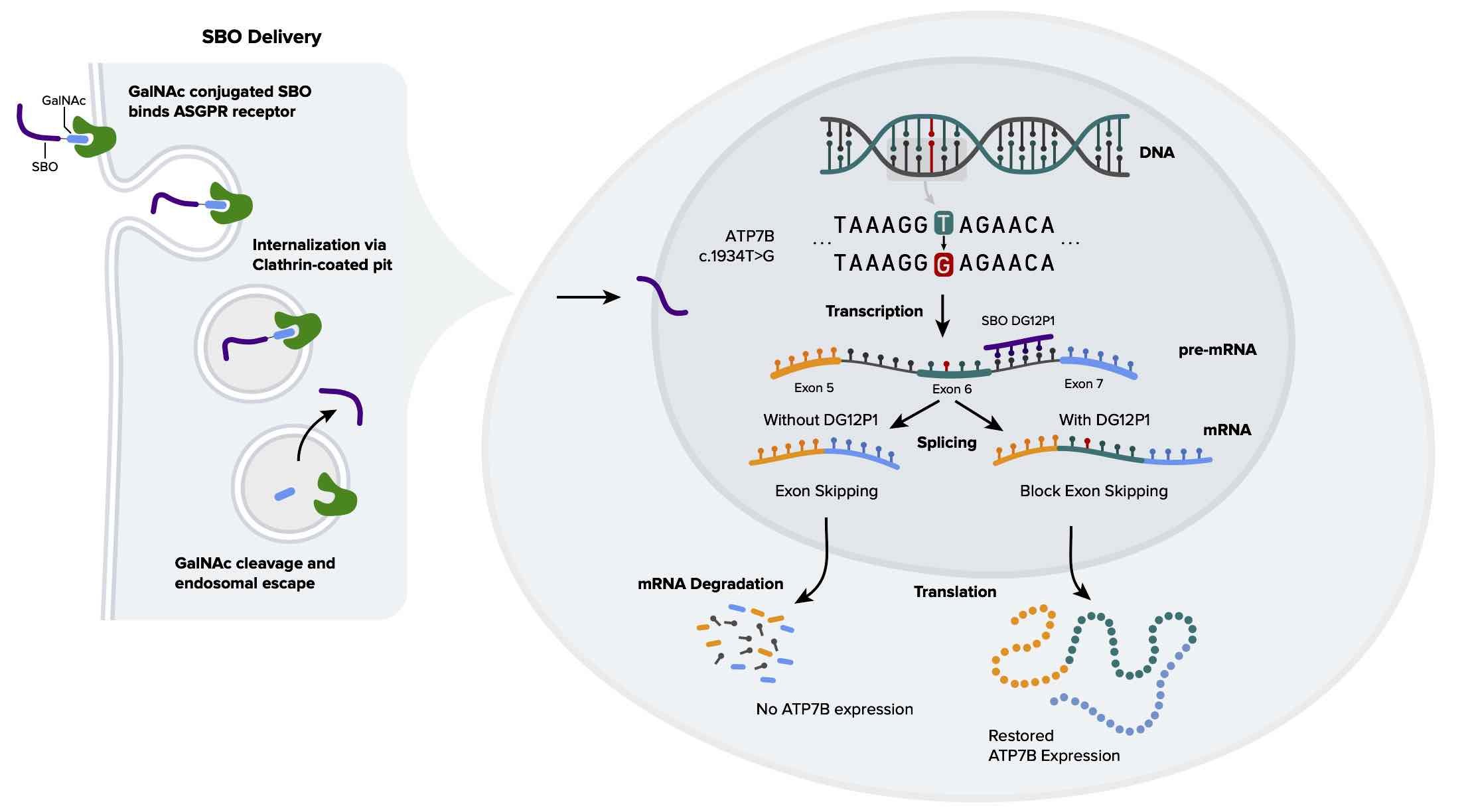
In early 2017, Deep Genomics developed a software system, called TargetRanch, to identify disease-causing genetic variants that can be remediated by therapeutic oligonucleotides (short stretches of chemically-modified DNA or RNA that attach to a specific place in the RNA). A key component of TargetRanch is artificial intelligence (AI) predictors trained on large-scale genomics datasets. TargetRanch scanned over 200,000 genetic variants implicated in rare disorders, and one of the best opportunities we found was a mutation in a gene called ATP7B, which causes Wilson Disease.
Wilson Disease is a rare disorder of copper homeostasis, which is present in about 20,000 individuals in North America and Europe, and is caused by loss of the copper transporter encoded by the ATP7B gene. Wilson Disease is fatal if untreated, and the current standard of care consists of drugs that actively remove copper or prevent its absorption in the gut. However, these drugs present significant adverse effects and adherence issues, and inadequate copper control, if protracted, can lead to severe neurological and liver damage later in life.
The variant we identified, c.1934T>G M645R, was first reported in 1997, but its mechanism was not understood. Since it directly changes the protein amino acid sequence (methionine (M) is replaced by arginine (R) at position 645), several researchers looked for alterations in protein stability, localization and function imparted by this specific change - but they could not observe any significant difference. Thanks to our AI predictors, we proposed for the first time that this variant is pathogenic because of a different type of effect, an RNA splicing alteration (RNA splicing is an intermediate processing step required to produce protein from DNA). Instructions for splicing are encoded in the RNA sequence, but they are too complex for humans to comprehend since they comprise hundreds, if not thousands, sequence motifs that often act in combination - hence the importance of AI predictors.
Follow-up experiments validated this prediction and demonstrated a profound reduction in ATP7B function (70%), indeed mediated by splicing alteration, which is documented in this peer-reviewed published paper.
Using the same AI predictive models, we then proceeded to design and experimentally test an oligonucleotide that restores correct splicing patterns and thus brings back the ATP7B functional level near the normal range. This is an exciting new approach, and one of the therapeutic oligonucleotides that Deep Genomics is developing for clinical trials.
"The experimental data was so clear in validating our AI model prediction - it was an exciting result to share back with the computational team." Carl Spickett, Senior Research Scientist in the Preclinical Research team.
"It was really gratifying to see our efforts in building the AI predictors culminate in this groundbreaking discovery." Shreshth Gandhi, Senior Research Scientist in the Predictive Systems team.
This is just one of the examples of the impact that our platform has had so far, and we are actively working on expanding the number and complexity of our therapeutic programs. If you’re also passionate about using software and technology to help cure genetically-defined diseases, let’s talk!